Artículo original
Biotecnología Vegetal Vol. 17, No. 1: 11 - 17, enero - marzo, 2017
Instituto de Biotecnología de las Plantas. UCLV. MES.
eISSN 2074-8647, RNPS: 2154
Identification and phylogeny of the tomato receptor-like proteins family
Identificación y filogenia de la familia de proteínas tipo receptoras en tomate
Ermis Yanes-Paz1**, Gioser María Ramos-Echazábal2*, Glay Chinea3, Yanelis Capdesuñer Ruiz4, Ramón Santos Bermúdez1
1Plant-Pathogen Interaction Laboratory, Universidad de Ciego de Ávila. Carretera Morón km 9. Ciego de Ávila. Ciego de Ávila. CP 69 450. Cuba. e-mail: eyanes@bioplantas.cu
2Department of Animal and Human Biology, Facultad de Biología, Universidad de La Habana. Havana 10400, Cuba.
3Biomedical Division, Center for Genetic Engineering and Biotechnology. PO Box 6162. La Habana. CP 10600. Cuba.
4Metabolic Engineering Lab, Universidad de Ciego de Ávila. Carretera Morón km 9. Ciego de Ávila. Ciego de Ávila. CP 69 450.
*These authors contributed equally to the manuscript
ABSTRACT
The receptor-like proteins (RLPs) play multiple roles in development and defense. In the current work 75 RLPs were identified in tomato (Solanum lycopersicum L.) using iterative BLAST searches and domain prediction. A phylogenetic tree including all the identified RLPs from tomato and some functionally characterized RLPs from other species was built to identify their putative homologues in tomato. We first tested whether C3-F-based phylogeny was a good indicator of functional relation between related proteins of different species. Indeed, the functionally characterized CLAVATA2 (CLV2), the maize ortholog FASCIATED EAR2 (FEA2) and a putative tomato CLV2 described in Uniprot clustered together, which validates the approach. Using this approach Solyc12g042760.1.1 was identified as the putative tomato homologue of TOO MANY MOUTHS (TMM). It was shown that proteins in the same cluster of the phylogenetic tree share functional relations since several clusters of functionally related proteins i.e. the Ve cluster, the Cf cluster, and the Eix clade were formed.
Keywords: phylogeny, receptors, RLP, tomato
Identificación y filogenia de la familia de proteínas tipo receptoras en tomate
RESUMEN
Las proteínas tipo receptoras juegan múltiples papeles en procesos de desarrollo y defensa. En este trabajo se identificaron 75 RLPs en tomate (Solanum lycopersicum L.) mediante el empleo de búsquedas BLAST iterativas y predicción de dominios. Se construyó un árbol filogenético que incluyó todas las RLPs identificadas en tomate y otras proteínas de este tipo caracterizadas funcionalmente en otras especies. Primero se comprobó si la filogenia basada en la región C3-F constituía un buen indicador de la relación funcional entre proteínas relacionadas de diferentes especies. Se comprobó que, de hecho, las proteínas CLAVATA2 y su ortólogo en maíz FEA2 (FASCIATED EAR2) y la probable CLAVATA2 de tomate descrita en Uniprot se agruparon en una misma rama, lo que valida el enfoque. Con este procedimiento se identificó Solyc12g042760.1.1 como el probable homólogo en tomate del gen TMM. Se demostró que las proteínas en el mismo cluster en el árbol comparten relaciones funcionales, pues se formaron varios cluster de proteínas relacionadas funcionalmente como por ejemplo el cluster Ve, el cluster Cf, y la clada Eix.
Palabras clave: filogenia, receptores, RLP, tomate
INTRODUCTION
The receptor-like proteins (RLPs) represent the second largest family of cell surface receptors in plants (Wang et al., 2010). Topologically, the eLRR-RLPs have an extracellular leucine-rich repeat domain, a transmembrane domain, and a short cytoplasmatic tail. The amino acid sequence of RLPs has been divided into the conserved domains A through G, where (A) is the putative signal peptide, (B) is the Cys-rich domain, (C) is the LRR domain, (D) is called the spacer, (E) is an acidic domain, (F) is the transmembrane domain, and (G) is a short cytoplasmic region. Furthermore, the LRR domain (C) is subdivided into three domains in which the non-LRR island C2 domain interrupts the C1 and C3 LRR regions (Jones and Jones, 1997).
Although the function of most eLRR-RLPs is still unknown, the functionally characterized members have been shown to play roles in the control of developmental processes and defense against pathogens (Wang et al., 2010). Characterized members of the tomato (Solanum lycopersicum L.) RLP gene family are the Cf genes which control resistance to various races of Cladosporium fulvum Cooke (Jones et al., 1994; Dixon et al., 1996; Thomas et al., 1997; Dixon et al., 1998; Takken et al., 1998), the Ve1 gene that confers resistance to Verticillium dahliae Klebahn and Verticillium albo-atrum Reinke & Berthold (Kawchuk et al., 2001; Fradin et al., 2009, Fradin et al., 2014). Other members include the tomato LeEix genes that recognizes an ethylene-inducing xylanase (EIX) of the biocontrol fungus Trichoderma viride Pers (Ron and Avni, 2004). RLPs implicated in the control of plant development include Arabidopsis TooManyMouths (TMM) that regulates stomatal distribution (Yang and Sack, 1995, Nadeau and Sack, 2002), and CLAVATA2 (CLV2) and its functional maize ortholog FEA2 that mediate meristem maintenance (Kayes and Clark, 1998; Jeong et al., 1999; Taguchi-Shiobara et al., 2001).
A total of 57 and 90 eLRR-RLP-encoding genes have been identified in Arabidopsis (Arabidopsis thaliana L.) and rice (Oryza sativa L.), respectively (Wang et al., 2008; Fritz-Laylin et al., 2005). In tomato, a previous study aimed at the identification of candidate pathogen recognition genes identified 176 LRR containing proteins. However, there are many proteins with LRR domains which do not fit the structure of RLPs. For that reason the real number of RLPs is still unknown in tomato. In this study, it was used a combination of iterative blast searches with protein domain analysis to identify all Cf9-like eLRR-RLPs in the tomato genome using the same criteria utilized for describing Arabidopsis RLPs (Fritz-Laylin et al., 2005). Also, it was used a phylogeny-based approach to predict homologs in tomato of previously characterized proteins in Arabidopsis and other species.
MATERIALS AND METHODS
Genome query
The Verticillium wilt disease resistance protein (Ve1) (UniProt code: C4NAS0) from S. lycopersicum was used to query the ITAG Release 2.3 (26-04-2011) SL2.40 database available at ITAG release 2.3 SL2.40 (SGN: http://solgenomics.net) for eLRR-RLPs. Iterative Blast searches (Altschul et al., 1997) were conducted using various members of this class.
Prediction of protein structures
Prediction of signal peptides (SP) and transmembrane domains (TM) was performed using Phobius (Käll et al 2004) (http://www.phobius.sbc.su.se), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and SignalP 3.0 (Emanuelsson et al., 2007) (http://www.cbs.dtu.dk/services/SignalP/) servers.
The identification of conserved domains in the proteins was performed using the batch search option in the Pfam database (Marshall et al 2004) (http://pfam.xfam.org/), the gathering threshold option was chosen. Additional manual checks were done using the Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) databases.
Phylogeny
The C3-F region was selected for the alignments and phylogeny reconstruction because all RLPs contain this region. Sequence alignments were performed using the MAFFT (Katoh et al., 2002) algorithm available in the webserver of the European Biotechnology Institute (http://www.ebi.ac.uk/Tools/msa/mafft/). The default parameters were used, except for the number of iterations which was increased to 20. The phylogenetic tree was built using the Neighbor Joining algorithm (Saitou and Nei, 1987) with 1000 bootstraps for branch support. The tree was drawn using the Mega version 5 (Tamura et al., 2011).
Identification of eLRR-RLPs in the tomato genome
To identify eLRR-RLP family members, an initial BLAST search was performed against the database of the International Tomato Genome Sequencing Project available at the Solgenomics (SGN) website (www.solgenomics.net) using the Ve1 (Fradin et al., 2009), a typical eLRR-RLP, as a query.
The E-value was set up to 1 to include sequences showing a relatively low similarity to Ve1. To saturate the query, iterative BLAST searches were performed using retrieved eLRR-RLP sequences with relatively low similarity to Ve1 as query. In each case, the E-value was set to 1 to retrieve all eLRR-RLPs from the tomato genome and iterative searches were performed until no new members were found. Subsequently, additional BLAST searches using sequences of previously characterized eLRR-RLPs (Hcr9-0 from S. lycopersicum (UniProt code: O49879; SGN: Solyc01g009690.1.1), LeEix1 (UniProt code: Q6JN47; SGN: Solyc01g009690.1.1) as queries were conducted.
This last step allowed verifying that the BLAST searches were saturated. To remove false positives, it was performed domain predictions were performed using batch search option of the Pfam database. Finally, it was manually it was checked the domain composition of every protein using the SMART and Pfam databases and the Phobius server followed by SignalP and TMHMM servers to check for signal sequences and transmembrane helixes, respectively. Since homology based methods can include LRR containing proteins which do not fit in the canonical structure of an eLRR-RLP, it was used the same stringent criteria used for Arabidopsis and rice RLP identification were used (Fritz-Laylin et al., 2005). Using these criteria 75 eLRR-RLPs were identified (Table 1).
Phylogeny of tomato eLRR-RLPs
In this study, the C3-F region was selected as the basis for phylogenetic tree reconstruction. Several characterized RLPs from different species were included in the study. We A classification did not follow theinto classification into GHG was not done, but adopted a similar procedure to define clades. It was defined a clade as subtrees containing at least two sequences, greater than 60% bootstrap support.
It was included tThe characterized RLPs from different species were included in the phylogenetic tree to determine the presence of putative homologs in tomato.
Prediction of tomato homologs of previously characterized RLPs in other species
The phylogeny based on the C3-F region placed together functionally characterized homologues involved in developmental processes of arabidopsis and maize
First, it was tested whether phylogeny based on C3-F was a good indicator of functional relation between related proteins of different species. However, there are no characterized orthologs in tomato of previously characterized RLPs from other species. Therefore, it was included the arabidopsis CLV2 and the maize (Zea mays L.) ortholog FEA2 were included in the analysis to test whether they cluster with putative tomato CLV2 which is reported as Uniprot accession F8WS88, but has not been functionally characterized.
RESULTS AND DISCUSSION
Identification of eLRR-RLPs in the tomato genome
Using the procedures described in Materials and methods, a total of 7 Using these criteriacriteria total of 75 eLRR-RLPs were identified (Table S1).
The included sequences typically encode a signal peptide (SP), an eLRR domain, a transmembrane segment (TM), and a short C-terminal tail. Instead of an HMM based on the structure of these proteins, it was used typical RLPs to query the databases and then manual analysis based on the domains structure of the proteins. Those proteins without a predicted TM were excluded (with a few exceptions based on the structure) to avoid including polygalacturonase inhibiting proteins (PGIPs) which are extracellular proteins with a similar structure to RLPs and other non Cf9-like proteins containing LRR domain. However, sequences without a predicted SP were included provided that they contain the typical domain structure of a Cf9-like RLP. Andolfo et al. (2013) described what they called “candidate pathogen recognition genes” in tomato which included proteins containing LRR domains such as the nucleotide-binding site (NBS), RLPs and the receptor-like kinases (RLK) gene families. Using this procedure they described 176 RLPs in tomato. However, many of these proteins do not fit the canonical structure of an RLP since they do not contain SP or transmembrane domain. Therefore, the numbers cannot be compared to the identified in Arabidopsis, rice or other species. If a comparison ofcomparison of the number of RLPs in tomato, rice and Arabidopsis is established, apparently the tomato genome contains almost three times more RLPs and twice as many RLPs as the arabidopsis and rice genomes, respectively. It was assume that this is Apparently, this is due to the fact that because different criteria were used to identify them. For that reason it was used similar criteria to identify the tomato RLPs as those used for arabidopsis and rice as described in Materials and Methods were used.
eLRR-RLPs comprise a variable group of proteins with low overall sequence similarity. Its sequence has been divided into seven different domains (Jones et al., 1994). Unlike RLKs which have been classified in subfamilies, there is not a classification in subfamilies for RLPs. However, in an attempt to group them, Fritz-Laylin et al. (2005) classified them into global homology groups (GHG) based on length and sequence similarity.
Of the 90 sequences in the tree, 70 are found in the 10 clades, the remaining 20 sequences were considered as phylogenetic singletons. Six clades contained only tomato proteins and three included proteins from different species. Arabidopsis AtRLP 30, 41 and 52 formed a separate cluster which did not include any tomato protein (Figure 1, S-1). Seven clades included at least one characterized member, the remaining three were tomato specific and did not include any functionally characterized protein.
The phylogeny based on the C3-F region placed together functionally characterized homologues involved in developmental processes of arabidopsis and maize
It was included both proteins in the phylogenetic tree and they formed a highly supported cluster (Figure 1, S-1) which also included the putative tomato CLV2 (Solyc04g056640.1.1). The fact that the two characterized orthologs from arabidopsis and maize clustered together could indicate that phylogeny based on the C3-F region may be used to predict the tomato homologs of functionally characterized proteins in other species On the other hand, Solyc04g056640.1.1 with 57% and 51% sequence identity with AtCLV2 and maize FEA2, respectively, is confirmed in this study as the putative tomato CLV2.
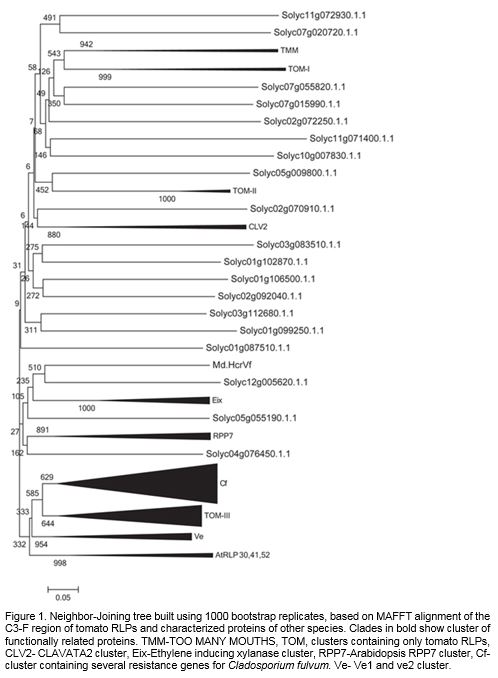
Figure 1. Neighbor-Joining tree built using 1000 bootstrap replicates, based on MAFFT alignment of the C3-F region of tomato RLPs and characterized proteins of other species. Clades in bold show cluster of functionally related proteins. TMM-TOO MANY MOUTHS, TOM, clusters containing only tomato RLPs, CLV2- CLAVATA2 cluster, Eix-Ethylene inducing xylanase cluster, RPP7-Arabidopsis RPP7 cluster, Cf- cluster containing several resistance genes for Cladosporium fulvum. Ve- Ve1 and ve2 cluster.
Prediction of tomato homologs of functionally characterized arabidopsis RLPs
eLRR-RLPs involved in developmental processes
Only two RLPs have been implicated in the control of developmental processes namely CLV2 (discussed above) and TMM which regulates proper stomata distribution by controlling the initiation of stomatal precursor cells (Yang and Sack, 1995; Nadeau and Sack, 2002). TMM closely clustered with Solyc12g042760.1.1 (52% of similarity) suggesting that it could be its homolog in tomato (Figure 1). Because of the central role of TMM in such an important process as stomatal cell fate determination (Nadeau and Sack, 2002) it is not a surprise that tomato contains a homologue of this gene which might also be present in the genomes of other plant species.
eLRR-RLPs involved in the response to specific pathogens
The Ve locus has been shown to comprise two genes, Ve1 and Ve2 (Kawchuk et al., 2001). In other species like cotton, resistance to this pathogen appears to be polygenic (Lüders et al., 2008). In the tomato genome these were identified as Solyc09g005090.1.1 and Solyc09g005080.1.1, with 100% identity to Ve1 and Ve2, respectively. Interestingly, Ve1 and Ve2 closely clustered with three other proteins of unknown function (Solyc01g098680.1.1, Solyc01g098690.1.1, Solyc10g076500.1.1) which share 43%, 49% and 48% identity with the functional Ve1 (Figure S1). It would be interesting to test whether these proteins play a role in defense. There are no Ve1 genes in Arabidopsis which is susceptible to Verticillium (Fradin et al., 2009).
The Cf proteins are the founding members of the eLRR-RLP family (Jones, 1994). Shortly after the characterization of Cf-9 (Jones et al., 1994), Cf-4, which maps to the same position, was cloned (Thomas et al., 1997). The Cf-4 gene from Solanum habrochaites S.Knapp & D.M.Spooner and the Cf-9 gene from S. pimpinellifolium L. map to allelic complex loci on the short arm of chromosome 1 (Jones et al., 1994;Thomas et al., 1997). Five homologues of S. pimpinelifolium Cf-9 have been described, namely Hcr9-9A, Hcr9-9B, Hcr9-9D, and Hcr9-9E. The corresponding locus in susceptible S. lycopersicum L. was denoted Hcr9-0 (Thomas et al., 1997). By analogy, the five homologues of Cf-4 have been designated Hcr9-4A, Hcr9-4B, Hcr9-4C, and Hcr9-4E (Thomas et al., 1997).
In addition, Cf-9 and Cf-4, Cf-2 and Cf-5 have been characterized in S. pimpinellifolium and the land race S. lycopersicum var. cerasiforme, respectively (Dixon et al., 1996; Dixon et al., 1998). These two latter genes map to the same position at a complex locus on chromosome 6 (Dickinson et al., 1993). Initially, three Cf-2 homologues namely Hcr2-2A, Cf-2.1 and Cf-2.2 were described (Dixon et al., 1996). However, a study on wild S. pimpinellifolium populations identified at least 26 different Cf-2 homologues (Caicedo and Schaal, 2004). For Cf-5, four homologues have been described, namely Hcr2-5A, Hcr2-5B, Cf-5 and Hcr2-5D (Dixon et al., 1998).
The ‘Heinz 1706-BG’ cultivar used for sequencing of the tomato genome apparently does not contain C. fulvum resistance genes since none homolog of the characterized Cf genes was found in the tomato genome sequence. However, the homolog of Hcr9-A from S. pimpinellifolium (UniProt code: O50028) designated as Hcr9-0 in S. lycopersicum (UniProt code: O49879) was identified as Solyc01g009690.1.1 98% identity with the reported sequence. At the Cf2/Cf5 where no functional homologs are expected in the Heinz 1706-BGgenome, the two putative non-functional homologs Hcr2-0A and Hcr2-0B were identified as Solyc06g008270.1.1 and Solyc06g008300.1.1 with 94 and 97% identity with the reported sequences. As expected, these four proteins form a solid cluster (97% bootstrap support).
Ron and Avni (2004) described a gene family corresponding to the LeEix locus of tomato. Two functional genes, LeEix1 and LeEix2, and likely pseudogene LeEix3 were identified and characterized as members of the eLRR-RLP family of proteins. LeEIX1, LeEIX2 and LeEIX3 were identified as Solyc07g008620.1.1, Solyc07g008630.1.1 and Solyc07g008640.1.1, respectively. They formed a highly supported cluster (Figure S1) which, interestingly, also includes the uncharacterized Solyc07g008590.1.1 and Solyc07g008600.1.1 which could probably have a related function.
CONCLUSIONS
According to the studystudy the tomato genome contains 75 RLP like genes. This work also shows that the use of a phylogeny based approach together with reciprocal BLAST and sequence identity comparisons is an effective method to predict homologues in tomato of functionally characterized genes in other species.
REFERENCES
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402
Caicedo AL, Schaal BA (2004) Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. Proceedings of the Natural Academic of Sciences of the United States 101 (50):17444–17449; doi: 10.1073/pnas.0407899101
Dickinson M, Jones DA, Jones JDG (1993) Close linkage between the Cf-2/Cf-5 and Mi resistance loci in tomato. Molecular Plant-Microbe Interaction 6 (3): 341–347
Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10 (11): 1915–1925
Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84 (3): 451–459
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nature Protocols 2 (4): 953-971; doi: 10.1038/nprot.2007.131
Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiology 156 (4): 2255–2265; doi: 10.1104/pp.111.180067
Fradin EF, Zhang Z, Ayala JCJ, Castroverde CDM, Nazar RN, Robb J, Liu CM and Thomma BPHJ (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiology 150 (1): 320-332; doi: 10.1104/pp.109.136762
Fradin EF, Zhang Z, Rovenich H, Song Y, Liebrand TW, Masini L (2014) Functional Analysis of the Tomato Immune Receptor Ve1 through Domain Swaps with Its Non-Functional Homolog Ve2. PloS One 9 (2): 1-14; doi: 10.1371/journal.pone.0088208
Fritz-Laylin LK, Krishnamurthy N, Tör M, Sjolander KV, Jones JD (2005) Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiology 138 (2): 611–62; doi: 10.1104/pp.104.054452
Jeong S, Trotochaud AE, Clark E (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 (10): 1925–1933
Jones DA, Jones JD (1997) The role of leucine-rich repeat proteins in plant defences. Advances in Botanical Research 24: 89–167; doi: 10.1016/S0065-2296(08)60072-5
Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266 (5186): 789–793
Käll L, Krogh A, Sonnhammer E (2004) A Combined Transmembrane Topology and Signal Peptide Prediction Method.Journal of Molecular Biology 338(5):1027-1036; doi: 10.1016/j.jmb.2004.03.016
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30 (14): 3059–3066
Kawchuk L, Hachey J, Lynch DR, Klcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R , Prufer D (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proceedings of the Natural Academic of Sciences of the United States 98 (11): 6511–6515; doi: 10.1073/pnas.091114198
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Method. Molecular Biology and Evolution 28 (10): 2731-2739; doi: 10.1093/molbev/msr121
Lüders RR, Galbieri R, Fuzatto MG, Cia E (2008) Inheritance of resistance to Verticillium wilt in cotton. Crop Breeding and Applied Biotechnology 8 (4): 265-270
Marshall M, Moxon S, Sonnhammer E L, Studholme DJ, Yeats C, Eddy S R (2004) The Pfam protein families database. Nucleic Acids Res 32: 138-141; doi: 10.1093/nar/gkh121
Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 (5573): 1697–1700; doi: 10.1126/science.1069596
Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16 (6): 1604–1615; doi: 10.1105/tpc.022475
Saitou N, Nei N (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4): 406-25
Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fascinated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes and Development 15 (20): 2755–2766; doi: 10.1101/gad.208501
Takken FLW, Schipper D, Nijkamp HJJ, Hille J (1998) Identification and Ds-tagged isolation of a new gene at the Cf-4 locus of tomato involved in disease resistance to Cladosporium fulvum race 5. Plant Journal 14 (4):401-411
Thomas CM, Jones DA, Parniske M, Harrison K, BalintKurti PJ, Hatzixanthis K, Jones JDG (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9 (12): 2209–2224
Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tor E, Zipfel C, de Wit PJGM, Jones JDG, Mahmut T, Thomma BPHJ (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiology 147 (2): 503–517; doi: 10.1104/pp.108.119487
Wang G, Fiers M, Ellendorff U, Wang Z, de Wit PJGM, Angenent G , Thomma BPHJ (2010) The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Critical Reviews in Plant Science 29 (5): 285–299; doi: 10.1080/07352689.2010.502082
Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 (12): 2227–2239; doi: 10.1105/tpc.7.12.2227
Recibido: 06-11-2016
Aceptado: 18-01-2017
Copyright (c) 2017 Biotecnología Vegetal
Biotecnología Vegetal eISSN 2074-8647, RNPS: 2154. ISSN 1609-1841, RNPS: 0397 Editada por: Instituto de Biotecnología de las Plantas. Universidad Central Marta Abreu de Las Villas. Carretera a Camajuaní km 5.5, Santa Clara, Villa Clara, Cuba CP 54 830 Tel: 53 42200124, e-mail: info@ibp.co.cu

Biotecnología Vegetal está bajo una Licencia Creative Commons Atribución-NoComercial 4.0 Internacional.
