Artículo original
Biotecnología Vegetal Vol. 18, No.1: 3 - 13, enero - marzo, 2018
Instituto de Biotecnología de las Plantas. UCLV. MES.
eISSN 2074-8647, RNPS: 2154
Bacillus megaterium: an endophytic bacteria from callus of Ilex paraguariensis with growth promotion activities
Bacillus megaterium: bacteria endofítica de callos de Ilex paraguariensis con actividad de promoción del crecimiento
Renata Lúcia Grunennvaldt¹, Juliana Degenhardt-Goldbach², Jéssica de Cássia Tomasi¹, Germana Davila Dos Santos³, Vânia Aparecida Vicente³, Cícero Deschamps¹
¹Crop Sciences Department, Federal University of Parana. Box:19061. Curitiba. PR. Brazil. CP 80035-050. e-mail: regrunennvaldt@gmail.com
²National Centre of Forestry Research. Colombo. PR. Brazil. CP 83411-000.
³Pathology Basic Department, Federal University of Parana. Box19031. Curitiba. PR. Brazil. CP 81.531-980.
ABSTRACT
Endophytic microorganisms live inside healthy plants and their isolation and identification may favor biotechnological applications. We isolated endophytic bacteria from Ilex paraguariensis callus cultivated in vitro for five months. Sequencing of 16S rRNA region to identify two isolates as Bacillus megaterium was used. The presence of bacterial cells in the intercellular spaces of callus cultures was detected by ultra structural analyses. The isolates were also evaluated for indole acetic acid (IAA) production and their potential in plant growth promotion of Phaseolus vulgaris L. The synthesis of IAA in the presence and absence of L-tryptophan was detected by colorimetric analyze for both isolates. In the presence of extracts from strains IPC06 and IPC07 the seedlings of P. vulgaris grew faster when compared to the control plants without the extract on in vitro tests.
Keywords: yerba mate, 16S rRNA, sequencing, tissue culture
RESUMEN
Los microorganismos endófitos viven dentro de plantas sanas y su aislamiento e identificación pueden favorecer las aplicaciones biotecnológicas. Los microorganismos endofíticos fueron encontrados en callos de Ilex paraguariensis cultivados in vitro durante cinco meses. Se usó la secuenciación de la región 16S rRNA para identificar dos aislados como Bacillus megaterium. La presencia de células bacterianas en los espacios intercelulares de los cultivos de callos se detectó mediante análisis ultraestructurales. Los aislamientos también se evaluaron para la producción de ácido indol acético (AIA) y su potencial en la promoción del crecimiento de las plántulas de Phaseolus vulgaris L. La síntesis de ácido AIA en presencia y ausencia de L-triptófano se detectó por análisis colorimétrico para ambos aislamientos. En presencia de extractos de las cepas IPC06 e IPC07, las plántulas de P. vulgaris crecieron más rápido en comparación con las plantas control sin el extracto en pruebas in vitro.
Palabras clave: yerba mate, 16S rRNA, secuenciación, cultivo de tejidos
INTRODUCTION
Ilex paraguariensis St. Hil (Aquifoliaceae), known as yerba mate, is a tree from the subtropical region of South America, present in southern Brazil, Paraguay, Uruguay and northern Argentina (Markowicz et al., 2007). It is used for preparing beverages (Bracesco et al., 2011), medicines and cosmetics, due to its stimulant and antioxidants properties, respectively.
Cloning by tissue culture techniques, yerba mate represents an important tool for genetic breeding of this species. Nevertheless, endophytic bacteria represents a problem, because after some subcultures under in vitro culture it kill the explants (Zaniolo and Zanette, 2002; Dutra et al., 2008; Luna et al., 2013. A recent study based on in vitro plants of this specie reveled a rich biota living inside the explants (Luna et al., 2013; Pérez et al., 2016).
Endophytic microorganism, existing in the inner tissues of living plants, have attracted increasing attention among taxonomists, ecologists, chemists and agronomists, mainly for affecting plant growth directly or indirectly. It is common sense, but these microorganisms represent an underexplored reservoir of novel species with potential interest for exploitation in pharmaceutical, agriculture and industry (Hallmann et al., 1997; Qin et al., 2011). In recent years, the plant-bacterial associations have received more attention due to their potential for biotechnological applications (Jin et al., 2014).
For this reason, the aims of this work was to isolate and identify endophytic bacteria from yerba mate in vitro callus and to evaluate their potential to produce auxin and promote plant growth.
MATERIALS AND METHODS
Plant material and callus induction
Plants of clone 6-156-6 from Ilex paraguariensis grown in the greenhouse, in EMBRAPA Forestry were used. The 2º/ 3º par of leaves were collected in antioxidant solution (0.5% ascorbic acid and 0.5% citric acid). The leaves were washed with neutral detergent in running tap water and disinfested as follow: 10 min in 1% (v/v) Cercobin®, 5 min in 0.05% (v/v) Hg2Cl2 and finally the leaves were rinsed three times in sterile distilled water. Afterwards, leaf discs with 2 cm diameter were cut and placed on Petri dishes containing 20 ml autoclaved (1.45 kg cm−2 for 20 min) medium.
The medium used for callus induction consisted of ¼ MS (Murashige and Skoog, 1962) salts and vitamins, 3% sucrose, 100 mg l-1 myo-inositol, 4.52 µM 2,4-dichlorophenoxyacetic acid (2,4-D), 4.52 µM zeatin with the pH adjusted to 5.8 prior to the addition of 0.7% agar. The Petri dishes containing the explants were incubated in the dark at 23 ± 2 ºC and subcultured every 30 days.
Isolation and identification of bacteria
Callus from clone 6-156-6, cultivated for 120 days, were placed on solid lysogeny broth (LB) medium (Sambrook et al., 1989) (10 g l -1 of NaCl, 10 g l -1 tryptone and 5 g l -1 yeast extract, pH 7.0) to allow the growth of endophytic bacteria. The Petri dishes were incubated for 3 days in the dark at 23 ± 2 ºC. Afterwards, bacteria growing around the explants were transferred to a fresh solid LB medium and cultivated for another 3 days at the same conditions. Then, single colonies were used for DNA extraction and sequencing (Quambusch et al., 2014).
The phylogenetic assignment of isolates was carried out by sequence analysis of the 16S rRNA gene. Total genomic DNA extraction was done as described by Vicente et al. (2008). Silica:celite (2:1) for maceration in CTAB (Cetyltrimethylammonium bromide) and CIA (chloroform:isoamyl alchol) for purification. After that, the DNA was quantified using NanoDrop 1000 (Spectrophotometer Thermo Scientific) and by agarose gel (1%) quantification. For the sequence analysis of the 16S rRNA gene, universal primers Y1f (5’TGGCTCAGAACGAACGCTGGCGGC3’) positions 20 to 43 and Y3r (5’TACCTTGTTACGACTTCACCCCAGTC3’) positions 1482 to 1507, which are specific for highly conserved regions of 16S rRNA, were used.
The PCR reactions were performed under the following conditions: PCR mixture (25 µl) containing 1.0 U Taq DNA polymerase, 2.0 mM MgCl2, 1x PCR buffer, 2.0 mM of each dNTP, 0.4 mM of each forward and reverse primer (Invitrogen, Brazil) and approximately 50 ng template DNA. It was submitted to 1 cycle of 94 ºC for 2 min, 35 cycles of 94 ºC for 1 min, 58 ºC for 1 min and 72 ºC for 2 min, 1 cycle of 72 ºC for 6 min. After, the amplicons integrity was observed on 1% agarose gel electrophoresis.
Amplicons were cleaned with exonuclease and shrimp alkaline phosphatase (sap) according to the instructions of the manufacturer. Amplicons were sequenced with bigdye terminator cycle sequencing Kit v. 3.1 (Applied Biosystems, Foster city, CA, USA) according to the instructions of manufacturer, and reaction mixtures were purified with sephadex g-50 fine (Ge health care Bio-sciences, Uppsala, Sweden). Sequences were analyzed on an ABI3130 DNA sequencer (Applied Biosystems).
Alignment and phylogenetic construction
Sequences were edited with the BioEdit software v.7.2.6 and compared to reference sequences at GenBank data sets (Supplement material). The alignment was performed with the MAFFT (Katoh and Standley, 2016), and visual inspection by the MEGA 7 version software (Kumar et al., 2016). The best evolutionary model to each dataset was estimated using the program MEGA v.7. The phylogenetic trees were constructed with 1000 bootstrap replicates using the Maximum Likelihood Implemented in Mega v7 software. New sequences generated in this study were deposited in NCBI GenBank nucleotide database (www.ncbi.nlm.nih.gov).
Ultra- structure analysis
I. paraguariensis callus with 5 months, where the growth of endophytic bacteria was visible under naked eye, were fixed in FAA as mentioned above and dehydrated in ethanol gradient series (70, 80, 90 and 100%, 10 min each step). The 100% ethanol step was repeated twice. After that, the explants were dried using the critical point technique (Baltec CPD 030) and coated with gold. The photomicrograph was taken by a JEOL (JSM 6360-LV) scanning electron microscope (JEOL Ltd, Tokyo, Japan) at the Electronic Microscopy Centre, UFPR.
Indole acetic acid (IAA) production
The ability of the isolates to produce IAA was determined qualitatively and quantitatively. The isolates were cultured overnight on LB medium at 28 °C, and then 25 µl of this bacterial culture were inoculated on a 50 ml Erlenmeyer flask containing 25 ml of LB medium with or without 0.1% tryptophan. The cultures were incubated for 72 h at 28 ºC at 125 rpm, in the dark. After that, the cultures were centrifuged at 6000 rpm for 30 min, and 2 ml of the supernatant was mixed with two drops of orthophosphoric acid and 4 ml of the reagent of Salkowski (50 ml, 35% perchloric acid, 1 ml 0.5M FeCl3) was added. The mixture was allowed to stand for 25 min. The indole acetic acid (IAA) production was measured qualitatively by the observation of a rose color and quantitatively only in the medium with tryptophan, by the measure of the intensity of the rose color at 530 nm using a spectrophotometer (UV Espectrophotometer 1800, Shimadzu). For determination, a standard curve (from 0-100 µg l-1) was also developed with a standard solution of pure indole-3-acetic acid. The experiment was conducted in triplicate.
Plant growth promotion
The isolates were cultivated separately in LB medium as described above with tryptophan to obtain the bacterial extract. The bacteria optical density (OD600) was determined and controlled to be between 0.8 and 1. The bacterial suspension was centrifuged and the supernatant was then autoclaved. Afterward 2.5% of bacterial extract was added to ¼ MS medium (Murashige and Skoog, 1962) before medium sterilization by autoclave. As control, LB medium without addition of the bacterial extract was used.
Seeds of Phaseolus vulgaris L. were disinfected using 1% (v/v) Cercobin for 10 min, and in 0.05% H2Cl2 for 5 min. Seeds were then rinsed three times with sterile distilled water and placed in 20 ml glass flasks containing the ¼ MS medium with or without addition of the bacterial extract. Three seeds per flasks with ten replicates were used. All flasks, lidded with plastic film, were incubated in a growth chamber at 23 ± 2 ºC under a 16 h photoperiod. After 20 days root and shoot lengths were measured and the dried weight was estimated (López-Bucio et al., 2007).
Statistical analysis
Data were analyzed with one-way analysis of variance (ANOVA) using the statistical program Assistat 7.7 and means were compared by the T- Student test at the 0.05 level of significance. For all the data, the homogeneity tests were performed by the Bartlett test.
RESULTS AND DISCUSSION
Isolation and identification of bacteria
Based on the sequence analysis of 16S rRNA region it was possible to identify two endophytic bacteria isolates (IPC06 and IPC07) from yerba mate callus as Bacillus megaterium clustered on clade very well supported (100%). The analysis of reference strains deposited in Genebank (http://www.ncbi.nlm.nih.gov/genbank/) revealed that from the total of 1288 sites evaluated for 16S region of RNAr, 923 were conserved, 336 variable, 201 were parsimoniously informative (Pi). The empirical base frequencies were pi (A): 0.25, pi(C):0.22, pi(G): 0.25 pi(T), 0.20, with 1000 bootstrap inferences using Paenibacillus polymyxa as outgroup.
The partial 16S rRNA gene sequence of the isolate showed 100% sequence similarity to B. megaterium strain CQN-25 (GeneBank accession no. KR347313.1) isolated from Apostichopus japonicas (Figure 1).
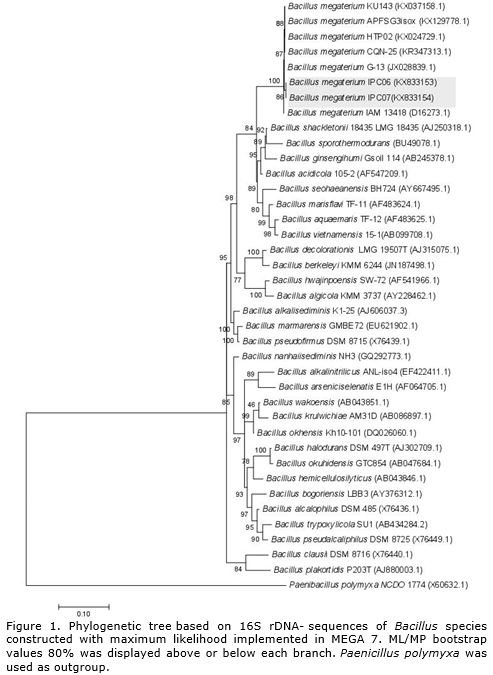
B. megaterium was first identified by Anton de Bary in 1884 as a soil Gram-positive bacteria (De Bary, 1884) and it is the largest of all bacilli (Korneli et al., 2013). B. megaterium is a spore-forming bacterium found in diverse habitats, mostly in soil, but also in seawater, sediment, rice paddies, honey, fish, and dried food (Vary et al., 2007) (Table 1). On the other hand, it has already been identified in several plant species as an endophytic bacteria in cotton (Gossypium spp.), sweet corn (Zea mays L.) (McInroy and Kloepper, 1995), coffee (Coffea sp.) (Nunes and De Melo, 2006), ginseng (Panax ginseng C.A.Mey.) (Vendan et al., 2010), ginger (Zingiber officinale Rosc.) (Chen et al., 2014) and black pepper (Piper nigrum L.) (Munjal et al., 2016).
In I. paraguariensis, B. megaterium was recently identified by Pérez et al. (2016), in leaves from plants grown under field conditions in Argentina. Besides the authors identified 20 taxa of endophytic bacteria from yerba mate leaves and stems. Another bacterium was also identified from I. paraguariensis during in vitro culture, Stenotrophomonas malthophilia (Luna et al., 2013). This bacterium was isolated from nodal segments of greenhouse-grown plants and hampered the I. paraguariensis in vitro propagation by bioreactor.
It was observed in preliminary tests, that the contaminations by endophytic microorganisms are lower in leaves when compared to stems on in vitro culture. In addition, it was observed the presence of endophytic bacteria only in a very few callus. The occurrence of bacteria in callus is also lower when compared with other works that used stems on in vitro culture from yerba mate (Zaniolo and Zanette, 2002; Dutra et al., 2008) (data not shown).
Pérez et al. (2016) observed the bacterial diversity in stems and leaves from I. paraguariensis, and identified 126 species in stems and 66 in leaves, confirming that the bacterial diversity is higher in stems than leaves from yerba mate. For this reason, our group is currently working to develop regeneration protocols for yerba mate based on regeneration of callus from in vitro leaves. Similar report was observed by Jin et al. (2014) in a medicinal plant, Stellera chamaejasme L. where the diversity of the bacterial community associated with stems was higher than that with leaves.
Ultra- structure analysis
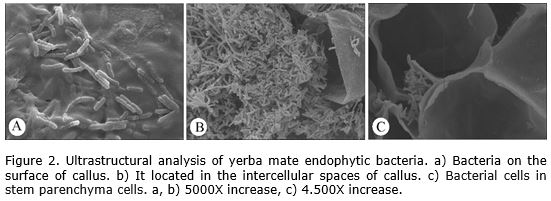
In the ultrastructural analysis it was possible to observe the presence of bacteria on the surface of callus (Figure 2 a). In addition, were also observed a high concentration of bacteria inside the cells (Figure 2 b) and in the intercellular spaces (Figure 2 c) that confirm the endophytic condition of the bacteria.
Endophytes were also observed colonizing other callus cultures. In Pinus sylvestris L. biofilms on the surface of bud-derived callus were detected using scanning electron microscopy (Pirttilä et al., 2002). Besides, callus from Piper spp., Taxus baccata subsp. wallichiana and Withania somnifera (L.) Dunal also showed the presence of endophytic bacteria (Kulkarni et al., 2007).
In the case of yerba mate, Luna et al. (2013) informed the presence of endophytic bacteria in nodal segments. By scanning electron microscopy the authors detected the presence of bacteria in the intercellular spaces of stem cortical parenchyma but no identified the species present. In another specie of the same genera, Ilex dumosa, previous report refer also in nodal segments cultivated in vitro the presence of endophytic bacteria (Luna et al., 2008). These reports confirm the presence of endophytic bacteria in tissue of Ilex plants and the possibility to find isolates with potential for different uses.
Indole acetic acid (IAA) production
Colorimetric analysis by the reagent of Salkowski of stationary phase cultures indicated that both isolates produce auxin in the presence and absence of L-tryptophan (Figure 3).
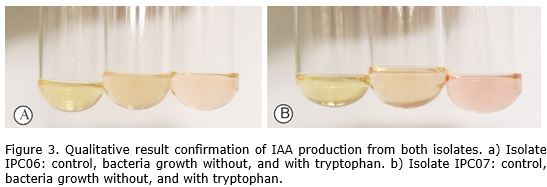
The reagent of Salkowski used in the colorimetric method allows the oxidation of indole-3-acetic acid (IAA) by peroxidase enzymes (Mayer, 1958). By using this reagent when the answer is positive for IAA production a pink color that varies from light pink to intense pink is obtained, depending on the concentration of the IAA (Meudt and Gaines, 1967). This method is an important option for qualitative and semi-qualitative determination of IAA that assure the presence of the hormone in the supernatant of bacterial cultures (Ambawade and Pathade, 2013).
The IAA synthesis occurs by different biochemical pathways: tryptophan-dependent pathways and tryptophan-independent pathway, given that, tryptophan has been identified as a main precursor for IAA biosynthesis pathways in bacteria (Spaepen et al., 2007). The production of IAA in the presence of L-tryptophan was 28.63 µg l-1 and 18.22 µg l-1 respectively for isolate IPC06 and isolate IPC07. The production of IAA in the absence of L-tryptophan was not evaluated quantitatively.
There are five proposed Tryptophan-dependent auxin biosynthesis pathways in bacteria: the indole-3-acetamide (IAM) pathway which is the best characterized pathway in bacteria, the tryptamine (TAM) pathway, the indole-3-acetonitrile (IAN) pathway and Trp side-chain oxidase (TSCO) pathway and the indole-3-pyruvic acid (IPyA) pathway, the major pathway for IAA biosynthesis in plants (Spaepen et al., 2007; Di et al., 2016). Besides these fives pathways in bacteria, there is one tryptophan-independent pathway branches from indole-3-glycerol phosphate or indole (Spaepen et al., 2007). However, the intermediate stages, and genes involved in tryptophan-independent pathways remain undefined (Fu et al., 2015). In this study, it was observed that the IAA production in both isolates from B. megaterium, occurred in the presence or absence of L-tryptophan, suggesting that this species can produce IAA by both pathways (Spaepen et al., 2007).
A bacterial tryptophan-independent pathway was demonstrated in other strains from B. megaterium: B. megaterium (AUX36) (Aziz et al., 2015), B. megaterium (MiR-4) (Ali et al., 2009) and other species from Bacillus: B. methylotrophicus (EB-26), B. subtilis (EB-04 and EB-55) and B. tequilensis (EB- 87) (Andrade et al., 2014). The activity of bacteria of the genus Bacillus is well documented, and several mechanisms have been suggested for the phytostimulatory activity of this group of plant growth promoting bacteria (PGPB). For this reason, the genus is considered very significant in agricultural applications (Ali et al., 2010).
Plant growth promotion assay
Statistical analysis of data revealed that the B. megaterium strains had significant effect on growth of P. vulgaris under in vitro conditions when compared with control (Table 1). Although similar results were observed for the two isolates tested (increase in shoot length and root length) here only present the results of strain IPC06. Similarity, increase in shoot and root fresh weight was observed. These results demonstrated that application of bacterial extract in culture medium favored the growth of bean seedlings (Figure 4).
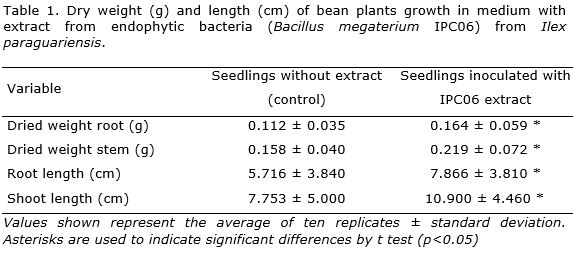
Regarding the root development, it was observed an increase in number and length of secondary roots and in the number of thin roots. It is well known that the division, expansion and differentiation of root cells and root elongation is influenced by IAA (Martinez-Viveros et al., 2010).

Other studies demonstrated the activity of bacteria in plant root formation. Similar response was reported by López-Bucio et al. (2007), where the inoculation of B. megaterium (UMCV1) in beans showed significant increase in the fresh and dry weights of plants. The plant-growth promotion was also related to modifications in root architecture. For Triticum aestivum L., the use of B. megaterium was reported to increase the root length, root number and root mass of the germinated seeds (Aziz et al., 2015).
Besides promoting the growth and development of the roots, the extract of B. megaterium accelerate the plants development when compared with the control. In addition to the IAA production, the endophytic bacteria may promote plant growth by a wide variety of mechanisms (Yu et al., 2016), including phosphate solubilization, nitrogen fixation, ammonia production and through the production of other phytohormones, such as cytokinins and gibberellic acids (Pereira et al., 2016).
The economic use of B. megaterium could be highly versatile. B. megaterium is known for plant growth promotion, its ability against plant pathogens and biocontrol (Munjal et al., 2016).
The success of B. megaterium use depends of plant and the isolate used. For example, in Arabidopsis it increased plant biomass, improved root system architecture, and augmented photosynthetic capacity (Zhou et al., 2016). For Vigna radiate (L.) Wilczek it caused significant increase in number of roots (Ali et al., 2009). In peppermint (Mentha piperita L.), plants inoculated with B. megaterium isolates manifested increase in plant height, number of leaves and branches, biomass and nutrient content, compared to non-inoculated plants (Sandeep et al., 2011). Nevertheless, in other plants such us Eucalyptus, the use of strain EUCB 26, with high production of IAA, had a negative effect on rooting, weight and length (Paz et al., 2012).
On the other hand, application of B. megaterium in tea (Camellia sinensis (L.) Kuntze) plants, promoted its growth, introduced the antagonistic activity to fungal pathogens and reduced sclerotial blight of tea caused by Sclerotium rolfsii. In addition, significant increases in phenolic compounds of tea plants were observed (Chakraborty et al., 2015).
According to the results, the endophytic bacterial strains from I. paraguariensis are promising microorganisms for promoting root and plant development.
CONCLUSION
Endophytic bacterial strains of B. megaterium isolated from callus of I. paraguariensis produce IAA and promote the growth of bean seedlings under in vitro conditions. This strains has potential as plant growth promoters.
ACKNOWLEDGMENTS
The authors thank Embrapa Forestry for providing laboratory facilities and plant material, the Laboratories of Biochemistry and of Electronic Microscopy Centre, Federal University of Parana, Brazil and CAPES for providing a grant to RLG.
REFERENCES
Ali B, Sabri AN, Hasnain KL, Hasnain S (2009) Quantification of indole-3-acetic acid from plant associated Bacillus spp. and their phytostimulatory effect on Vigna radiata (L.). World Journal of Microbiolgy Biotechnology 25 (1):519–526; doi: 10.1007/s11274-008-9918-9
Ambawade MS, Pathade GR (2013) Production of indole acetic acid (IAA) by Stenotrophomonas maltophilia BE25 isolated from roots of banana (Musa spp). International Journal of Science and Research 4(1): 2644–2650
Andrade LF, Souza GLOD de, Nietsche S, Xavier AA, Costa MR, Cardoso AMS, Pereira M CT, Pereira DFGS (2014) Analysis of the abilities of endophytic bacteria associated with banana tree roots to promote plant growth. Journal of Microbiology 52 (1): 27–34; doi: 10.1007/s12275-014-3019-2
Aziz K, Nawaz M, Nazir J, Anjum AA, Yaqub T, Ahmad MUD, Rehman MU, Aziz G, Khan M (2015) Isolation, characterization and effect of auxin producing bacteria on growth of Triticum aestivum. The Journal of Animal & Plant Sciences 25(4): 1003–1007
Bracesco N, Sanchez A G, Contreras V, Menini T, Gugliucci A (2011) Recent advances on Ilex paraguariensis research: minireview. Journal of ethnopharmacology 136(3): 378–384
Chakraborty AP, Chakraborty BN, Chakraborty U (2015) Bacillus megaterium from tea rhizosphere promotes growth and induces systemic resistance in tea against Sclerotium rolfsii. Indian Phytopathology 68(3): 237–247
Chen T, Chen Z, Ma GH, Du BH, Shen B, Ding YQ, Xu K (2014) Diversity and potential application of endophytic bacteria in ginger. Genetics and Molecular Research 13(3): 4918–493; doi: 10.4238/2014.July.4.6
De Bary A (1884) Vergleichende morphologie und biologie der pilze mycetozoen und Bacterien. Wilhelm Engelman
Di DW, Zhang C, Luo P, An CW, Guo GQ (2016) The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regulation 78(3): 275–285; doi: 10.1007/s10725-015-0103-5
Dutra LF, Hansel FA, Wendling I (2008) Introdução ao cultivo in vitro de Erva-mate (Ilex paraguariensis). Boletim de Pesquisa e Desenvolvimento 61(1): 1–33
Fu S-F, Wei J-Y, Chen H-W, Liu YY, Lu HY, Chou JY (2015) Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant signaling & behavior 10(8) e1048052; doi: 10.1080/15592324.2015.1048052
Hallmann J, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Canadian Science Publishing 43(1): 895–914
Jin H, Yang XY, Yan ZQ, Liu Q, Li XZ, Chen JX, Zhang DH, Zeng LM, Qin B (2014) Characterization of rhizosphere and endophytic bacterial communities from leaves, stems and roots of medicinal Stellera chamaejasme L. Systematic and Applied Microbiology 37(5): 376–385; doi: 10.1016/j.syapm.2014.05.001
Katoh K, Standley DM (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32(13): 1933–1942; doi: 10.1093/bioinformatics/btw108
Korneli C, David F, Biedendieck R, Jahn D, Wittmann C (2013) Getting the big beast to work-Systems biotechnology of Bacillus megaterium for novel high-value proteins. Journal of Biotechnology 163(2): 87–96; doi: 10.1016/j.jbiotec.2012.06.018
Kulkarni AA, Kelkar SM, Watve MG, Krishnamurthy K V (2007) Characterization and control of endophytic bacterial contaminants in in vitro cultures of Piper spp., Taxus baccata subsp. wallichiana, and Withania somnifera. Canadian Journal of Microbiology 53(1): 63–74; doi: 10.1139/w06-106
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33(7): 1870–1874; doi: 10.1093/molbev/msw054
López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E (2007) Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an uxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Molecular Plant-Microbe Interactions 20(2): 207–217
Luna C, Acevedo R, Collavino M, González A, Mroginski L, Sansberro P (2013) Endophytic bacteria from Ilex paraguariensis shoot cultures: Localization, characterization, and response to isothiazolone biocides. In Vitro Cellular and Developmental Biology - Plant 49(3): 326–332; doi: 10.1007/s11627-013-9500-5
Luna C, Collavino M, Mroginski L, Sansberro P (2008) Identification and control of bacterial contaminants from Ilex dumosa nodal segments culture in a temporal immersion bioreactor system using 16S rDNA analysis. Plant Cell, Tissue and Organ Culture 95(1): 13–19; doi: 10.1007/s11240-008-9408-7
Markowicz D, Moura de Oliveira D, Teixeira-Matsumoto RL, de Oliveira-Carvalho P, Ribeiro ML (2007) Yerbamate: pharmacological properties, research and biotechnology. Medicinal and Aromatic Plant Science and Biotechnology 1(1): 37–46
Martinez-Viveros O, Jorquera MA, Crowley DE, Gajardo G, Mora ML (2010) Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. Journal of Soil Science and Plant Nutrition 10(3): 293–319
Mayer AM (1958) Determination of indole acetic acid by the Salkowsky reaction. Nature 162(1): 1670–1671
McInroy JA, Kloepper JW (1995) Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant and Soil 173(2): 337–342; doi: 10.1007/BF00011472
Meudt WJ, Gaines TP (1967) Colorimetric Determination. Plant Physiology 42(1): 1395–1399
Munjal V, Valiya A, Sheoran N, Kundu A, Venugopal V, Subaharan K, Rajamma S, Eapen S J, Kumar A (2016) Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium. Biological Control 92(1): 66–76; doi: 10.1016/j.biocontrol.2015.09.005
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioessays with tobacco tissue cultures. Physiology Plantarum 15: 473–479
Nunes F V, De Melo IS (2006) Isolation and characterization of endophytic bacteria of coffee plants and their potential in caffeine degradation. Environmental Toxicology 10(1): 293–297; doi: 10.2495/ETOX060291
Paz ICP, Santin RCM, Guimarães AM, Rosa OPP (2012) Eucalyptus growth promotion by endophytic Bacillus spp. Genetics and Molecular Research 11(4): 3711–3720
Pereira SIA, Monteiro C, Vega AL, Castro PML (2016) Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecological Engineering 87(1): 91–97; doi: 10.1016/j.ecoleng.2015.11.033
Pérez ML, Collavino MM, Sansberro PA, Mroginski LA, Galdeano E (2016) Diversity of endophytic fungal and bacterial communities in Ilex paraguariensis grown under field conditions. World Journal of Microbiology and Biotechnology 32(4): 1–15; doi: 10.1007/s11274-016-2016-5
Pirttilä AM, Laukkanen H, Hohtola A (2002) Chitinase production in pine callus (Pinus sylvestris L.): A defense reaction against endophytes? Planta 214(6): 848–852; doi: 10.1007/s00425-001-0709-x
Qin S, Xing K, Jiang J, Xu L (2011) Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Applied Microbiology and Biotechnology 89(3): 457–473; doi: 10.1007/s00253-010-2923-6
Quambusch M, Pirttilä AM, Tejesvi M V, Winkelmann T, Bartsch M (2014) Endophytic bacteria in plant tissue culture: Differences between easy and difficult to propagate Prunus avium genotypes. Tree Physiology 34(5): 524–533; doi: 10.1093/treephys/tpu027
Sambrook J, Fritsch EF, Maniatis T, Spring HLC (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York; ISBN: 0-87969-309-6
Sandeep C, Raman RV, M R (2011) Effect of inoculation of Bacillus megaterium isolates on growth, biomass and nutrient content of Peppermint. J Phytol 3(11): 19–24
Spaepen S, Vanderleyden J, Remans R, Patra S, Gowda TKSC and Mulla SR (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiology Reviews 31(4): 425–448; doi: 10.1111/j.1574-6976.2007.00072.x
Vary PS, Biedendieck R, Fuerch T, Meinhardt F, Rohde M, Deckwer WD, Jahn D (2007) Bacillus megaterium from simple soil bacterium to industrial protein production host. Applied Microbiology and Biotechnology 76(5): 957–967; doi: 10.1007/s00253-007-1089-3
Vendan RT, Yu YJ, Lee SH, Rhee YH (2010) Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. Journal of Microbiology 48(5): 559–565; doi: 10.1007/s12275-010-0082-1
Vicente VA, Attili-Angelis D, Pie MR, Queiroz-Telles F, Cruz LM, Najafzadeh MJ, Hoog GS de, Zhao J, Pizzirani-Kleiner A (2008) Environmental isolation of black yeast-like fungi involved in human infection. Studies in Mycology 67(1): 137–144
Yu J, Yu ZH, Fan GQ, Wang GH, Liu XB (2016) Isolation and characterization of indole acetic acid producing root endophytic bacteria and their potential for promoting crop growth. Journal of Agricultural Science and Technology 18(1): 1381–1391
Zaniolo SR, Zanette F (2002) Micropropagação de erva-mate. Scientia Agraria 1(1): 1-6
Zhou C, Ma Z, Zhu L, Xiao X, Xie Y, Zhu J (2016) Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. International Journal of Molecular Sciences 17(976): 1–18; doi: 10.3390/ijms17060976
Recibido:15-11-2017
Aceptado:30-01-2018
Copyright (c) 2018 Biotecnología Vegetal
Biotecnología Vegetal eISSN 2074-8647, RNPS: 2154. ISSN 1609-1841, RNPS: 0397 Editada por: Instituto de Biotecnología de las Plantas. Universidad Central Marta Abreu de Las Villas. Carretera a Camajuaní km 5.5, Santa Clara, Villa Clara, Cuba CP 54 830 Tel: 53 42200124, e-mail: info@ibp.co.cu

Biotecnología Vegetal está bajo una Licencia Creative Commons Atribución-NoComercial 4.0 Internacional.
